With over 20+ years in Enterprise Storage, High-Speed Networking, Advanced Computation performance engineering, and later leveraging these platforms for Healthcare and Life Sciences research, a different architecture is needed from the ground up. Today's enterprise storage has primarily adapted to accommodate the needs of industry sector businesses for emerging technologies. The requirements for full-stack AI Solutions for Large Language Models (LLMs) and Generative AI workloads quickly break the current performance limits of the highest-performing enterprise storage arrays, high-speed networking, and advanced computation.
During my post-graduate research at UC Berkeley, I combined my professional background in enterprise storage, high-speed networking, and advanced computation performance engineering with a passion project I started in 2012, leveraging these related platforms to help with pediatric diseases and treatment research. During my UC Berkeley Project, I developed an AWS cloud-based solution that utilized baseline genomic sequencing data and pediatric patient genomic sequencing data to create Variant Call Format (VCF) Files for the applicable chromosomes for Medulloblastoma precision medicine approaches, the most common form of pediatric brain cancer. The VAST Data Platform and Supermicro Hyper-Scale Servers with NVIDIA BlueField-3 DPUs are promising because this solution accelerates innovation possibilities to levels that have never been possible. We could only dream of these types of technological capabilities. Healthcare and Life Sciences capabilities are paving the way to save lives through precision medicine treatments and drug development in a constantly evolving ecosystem. This pathway enables more accurate patient treatments, leading to significant advancements in remission outcomes and an unprecedented evolution in disease treatment methods. Simultaneously, it drastically reduces treatment costs. This example of Healthcare and Life Sciences is only one use case example. VAST Data Platform and Supermicro Hyper-Scale Servers with NVIDIA BlueField-3 DPUs unlock and enable these enormous possibilities in every business/industry sector.
To illustrate the massive capabilities of this solution architecture, we approach six main sections/areas of focus outlined below.
Section 1 - We'll cover the AWS Genomics Secondary Analysis for Precision Medicine Approaches solution that I created during my UC Berkeley research to provide the foundational background of what we are trying to accomplish with pediatric precision medicine in this area of focus example.
Section 2 - We'll cover the VAST Data Platform, including VAST DataStore, DataBase, DataSpace, and DataEngine, and how the VAST Data Platform is fundamentally different. We'll also explore how traditional enterprise storage, which was not initially designed for LLMs and Generative AI workloads, presents challenges.
Section 3 - We'll leverage the AWS Genomics Secondary Analysis for Precision Medicine Approaches solution I created but fully utilize the VAST Data Platform.
Section 4 - We'll dive into the VAST Data Platform and NVIDIA BlueField-3 DPUs for massive storage acceleration.
Section 5 - We'll discuss how the VAST Data Platform with NVIDIA BlueField-3 DPUs can be utilized for Pediatric Disease Precision Medicine Approach Acceleration.
Section 6 - To wrap up, we'll discuss how VAST Data Storage and Supermicro with NVIDIA BlueField-3 DPUs can be leveraged for Advanced Healthcare and Life Sciences Research Architecture for Pediatric Cancer Precision Medicine Research.
As someone who has spent over two decades focusing on enterprise storage performance engineering, I am beyond impressed with the massive potential of the VAST Data Platform. To meet the demanding needs of the most sophisticated AI Workloads, a full-stack AI Solution for LLM and Generative AI workloads is mandatory to scale, and VAST Data Storage and Supermicro with NVIDIA BlueField-3 DPUs unlock endless potential and possibilities. The capabilities of the future are here now!
Section 1 - AWS Genomics Secondary Analysis for Precision Medicine Approaches Overview
Genomics Secondary Analysis for Precision Medicine Approach
Below is a detailed walkthrough of the process, from a basic overview of genomic sequencing to leveraging genomic data for precision treatments from an AWS Cloud Solutions perspective.
Through the above video and presentation, we begin with a thorough understanding of Genomics Sequencing and a step-by-step guide on uploading fastq.gz Sequence Files into an AWS S3 Bucket. We then delve into creating Docker Images for BWAMem, SAMTools, and BCFTools and uploading these to an AWS S3 Bucket.
Subsequently, we demonstrate pushing these Docker Images to AWS Elastic Container Registry (ECR) Repositories for easy access and deployment. We then transition into creating the AWS Fargate Compute Environment, an essential platform for managing and running Docker containers.
Next, we will create an AWS Batch Queue utilizing the AWS Fargate Compute Environment. The focus on the AWS Batch Queue is followed by an explanation of creating AWS Batch Job Definitions, a crucial step in defining how jobs will be run.
Following this, we introduce the AWS Step Functions State Machine, outlining its workflow and demonstrating how to start execution and output to AWS S3.
Finally, we apply all these concepts to a real-world example - leveraging genomic data for the precision treatment of Pediatric Medulloblastoma (the most common form of pediatric brain cancer).
VCF Data Pipeline Into Relational and Non-Relational Databases and Machine Learning Approach
The Genomics Secondary Analysis Pipeline approach can further streamline the storage and analysis of VCF files within a database environment by ingesting outputs into AWS Relational Database Service (RDS) or AWS DynamoDB. For example, following the genomic analysis of children with Cystic Fibrosis, the VCF files containing complex variant data can be parsed and loaded into RDS or DynamoDB, easing the querying and analysis tasks. This supports advanced queries that can correlate specific genetic mutations with phenotypic data.
Utilizing an innovative solution combining CRISPR technology, scientists could further develop precise disease models by editing the genomes of cellular models to reflect mutations found in Cystic Fibrosis. Alternately, non-CRISPR techniques, like RNA interference or antisense oligonucleotides, allow researchers to modulate gene expression to understand the disease pathways further and identify new drug targets.
In drug discovery, AWS SageMaker can significantly enhance the screening process by analyzing vast libraries of chemical compounds, potentially pinpointing drugs that can be repurposed as gene therapies for Cystic Fibrosis. Machine Learning models, trained on existing pharmacological data, could predict how new or existing compounds might interact with mutated genes or proteins involved in the disease.
High-Level Workflow (Below)
fastq.gz to S3 ->
Docker Images (BWAMem, SAMTools, and BCFTools) Creation and to S3 ->
Docker Images from S3 to ECR ->
Docker Images from ECR to Fargate Compute Environment ->
Batch Queue Creation Utilizing the AWS Fargate Compute Environment ->
Batch Job Definitions ->
Step Functions State Machine Workflow Creation ->
Start Step Functions State Machine Workflow to Produce Variant Call Format Files (VCF) to S3 ->
Variant Call Format Files (VCF) to AWS Relational Database Service (RDS) or AWS DynamoDB ->
Leveraging AWS SageMaker for Machine Learning Model Development
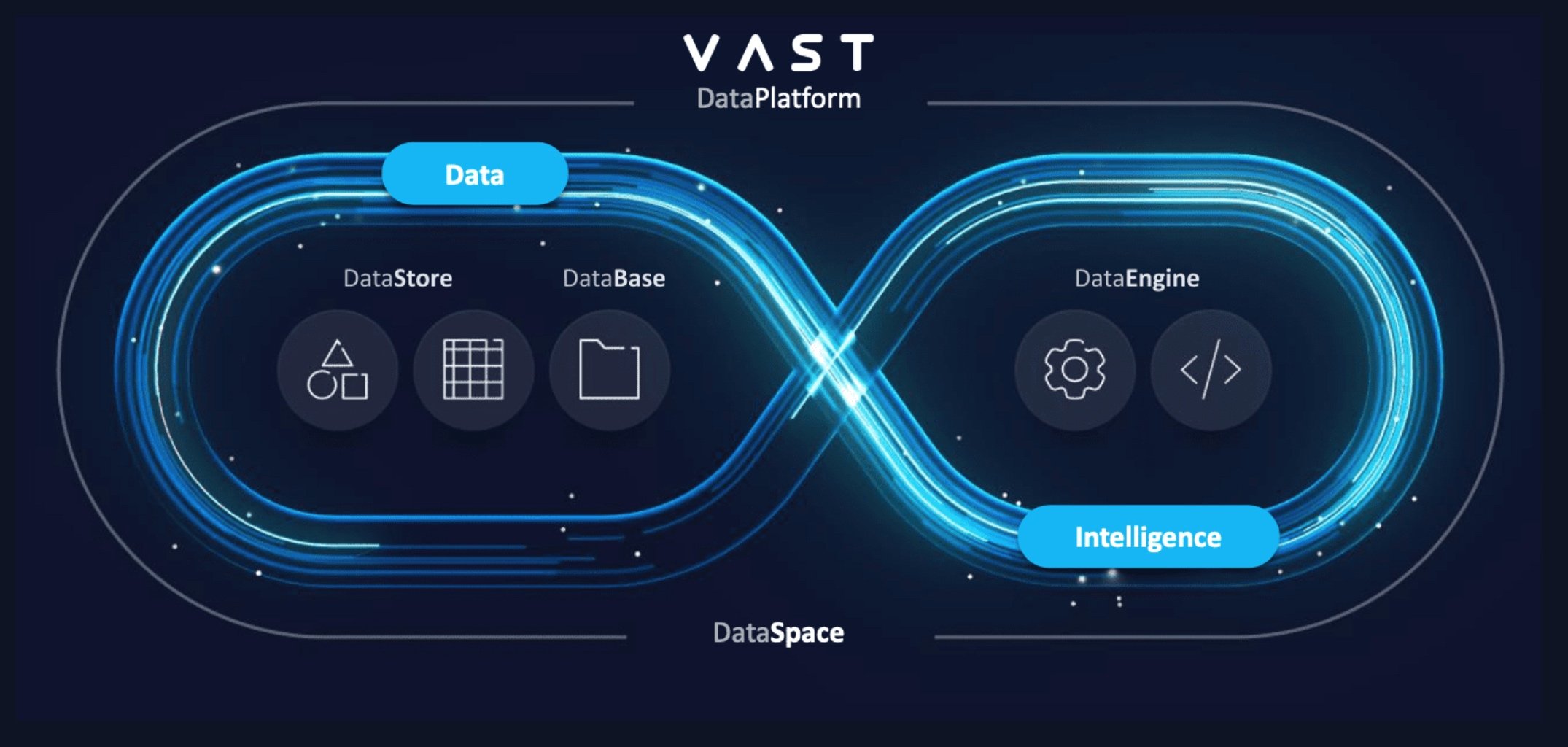
Section 2 - The VAST Data Platform
With its DataStore component at the forefront, the VAST Data Platform represents a paradigm shift in data storage and management. VAST DataStore is particularly tailored to meet the demanding requirements of LLMs and Generative AI workloads. Unlike traditional enterprise storage architectures that often struggle with scalability, performance consistency, and adaptability to modern data needs, the VAST Data Platform addresses these challenges head-on through innovative architectural decisions, performance optimizations, and a comprehensive suite of features.
Addressing Traditional Enterprise Storage Pitfalls
Traditional enterprise storage systems frequently encounter limitations due to their rigid architecture, which can lead to inefficiencies in handling the dynamic and large-scale data requirements of LLMs and Generative AI applications. These systems often struggle with the following:
Scalability: Difficulty scaling storage and computing independently, leading to resource overprovisioning or bottlenecks.
Performance: Inconsistent performance, particularly with high concurrency or large-scale data access, impacting AI model training and inference times.
Data Management: Inefficient data management and access mechanisms that complicate the integration and processing of diverse data types.
VAST DataStore: A Foundation for AI Workloads
The VAST DataStore, as part of the VAST Data Platform, directly addresses these challenges through:
Disaggregated, Shared-Everything (DASE) Architecture: This innovative approach allows independent scaling of compute and storage resources, ensuring that AI workloads can be dynamically supported without the inefficiencies associated with traditional storage systems. The DASE architecture also facilitates low-latency access to data, which is crucial for the performance-sensitive nature of AI applications.
Exceptional Snapshot Capabilities: Supporting up to 1 million snapshots per cluster, VAST DataStore ensures robust data integrity and recovery options. This feature is vital for AI workloads, where data consistency and the ability to revert to specific points in time can significantly impact model training and development.
Advanced Use of NVMe SSDs: VAST DataStore leverages NVMe SSDs for storage and offers high-speed data access, dramatically reducing the latency and throughput issues commonly encountered in AI and ML workloads. This technology guarantees seamless progress in data-intensive operations, like training large models, without delays.
Universal Access: The platform's shared-everything model simplifies data management and enhances performance by providing direct, low-latency paths to the storage infrastructure. This feature is particularly beneficial for distributed AI applications requiring efficient data sharing and access across multiple nodes.
Beyond Storage: Comprehensive Support for AI Workloads
The VAST Data Platform extends its capabilities beyond mere storage, offering specialized components designed to meet the multifaceted needs of AI applications:
VAST DataBase: Tailored for handling structured data, VAST DataBase supports high-performance queries and analytics, which is essential for AI models that rely on structured datasets for training and inference.
VAST DataSpace: This component focuses on decentralized data management, enabling seamless collaboration and data sharing across distributed environments. It is particularly useful for AI workloads spanning multiple locations, ensuring data consistency and accessibility.
VAST DataEngine: Designed to accelerate data processing tasks, VAST DataEngine optimizes the performance of data-intensive AI operations, such as preprocessing and transformation. This capability is crucial for preparing extensive AI model training and deployment datasets.
By integrating these components into a unified platform, the VAST Data Platform not only overcomes the limitations of traditional storage architectures but also provides a comprehensive solution that addresses the end-to-end needs of LLMs and Generative AI workloads. From high-performance storage and advanced data management to specialized support for structured and unstructured data, the platform is uniquely positioned to empower organizations to harness the full potential of their AI initiatives.
-Resources
Section 3 - VAST Data Platform Genomics Secondary Analysis for Precision Medicine Approaches Overview
Creating a genomics solution on the VAST Data Platform, tailored explicitly for producing Variant Call Format (VCF) files for precision medicine applications such as Pediatric Medulloblastoma and Cystic Fibrosis, involves leveraging the comprehensive suite of services offered by VAST DataStore, DataBase, DataSpace, and DataEngine. Below is an approach to leverage my previous AWS Genomics Secondary Analysis for Precision Medicine Approach with the VAST Data Platform to accelerate the solution with significant energy savings.
VAST Data Platform Ecosystem (Ref 1)
1. Data Ingestion and Storage
VAST DataStore: Begin by ingesting raw genomics sequencing data files (e.g., fastq.gz) into the VAST DataStore. Given its high performance and scalability, the DataStore can handle the large volumes of data typically associated with genomics sequencing. NVMe SSDs ensure rapid data access and transfer rates, which are crucial for time-sensitive genomics analyses.
2. Data Preparation and Processing
VAST DataBase & DataEngine: Utilize VAST DataBase to organize and manage the sequencing data. The DataBase's ability to handle massive datasets efficiently makes it ideal for genomics workloads. For the processing steps, such as alignment and variant calling, leverage VAST DataEngine to deploy containerized bioinformatics tools like BWAMem, SAMTools, and BCFTools. The DataEngine supports containerized environments, facilitating the creation, management, and execution of these bioinformatics workflows at scale.
3. Scalable Computing Environment
VAST DataSpace: With significant computational resources to process genomics data, VAST DataSpace can provide a scalable computing environment. It allows for efficiently running containerized bioinformatics tools across a distributed infrastructure, ensuring that resource-intensive tasks are completed swiftly. This is analogous to utilizing AWS Fargate in cloud environments but is seamlessly integrated within the VAST platform for on-premises deployments.
4. Workflow Management and Execution
Utilize the orchestration capabilities within VAST DataSpace to manage the workflow from raw sequence data to the generation of VCF files. This includes automating the sequence of processing steps—alignment, sorting, and variant calling—to produce VCF files efficiently. The orchestration tool would manage dependencies between tasks, handle task scheduling on available compute resources, and retry failed tasks as necessary.
5. Data Analysis and Storage for Precision Medicine
Once VCF files are generated, store them in VAST DataStore for long-term retention and accessibility. To further analyze and support precision medicine applications, integrate the VCF data with patient phenotypic data stored in VAST DataBase. The DataBase's performance capabilities are crucial for running complex queries across genomic and clinical data sets to identify relevant genetic variants associated with conditions like Pediatric Medulloblastoma and Cystic Fibrosis.
6. Leveraging Advanced Analytics and Machine Learning
For advanced analytics and Machine Learning purposes, including predictive modeling and identifying potential treatment paths, utilize VAST DataEngine to integrate with Machine Learning platforms and tools. This could involve training models on genomic and clinical data to predict disease outcomes or treatment responses.
Organizations can create a comprehensive and scalable genomics data analysis platform by leveraging the integrated capabilities of VAST DataStore, DataBase, DataSpace, and DataEngine. This platform would facilitate the rapid processing of genomics data to produce VCF files and support the broader objectives of precision medicine by enabling advanced analytics and insights into disease mechanisms and treatment responses.
High-Level Workflow (Below)
fastq.gz to VAST DataStore ->
VAST DataBase -> Organize and Manage the Sequencing Data ->
DataEngine -> Deploy Containerized Bioinformatics Tools (BWAMem, SAMTools, and BCFTools) ->
VAST DataSpace -> Manage Workflow from Raw Sequence Data to the Generation of VCF Files) ->
VAST DataBase -> Storage of VCF Data with Patient Phenotypic Data ->
VAST DataEngine -> Integrate with ML Platforms and Tools (Example: Databricks, TensorFlow, etc.) (Training Models on Genomic Data Combined with Clinical Data to Predict Disease Outcomes or Response to Treatments)
Section 4 - VAST Data Platform and NVIDIA BlueField-3 DPUs for Massive Storage Acceleration
NVIDIA BlueField-3 DPU (Ref 2)
Modern storage ecosystems necessitate robust networking capabilities and the flexibility to support various storage software functionalities, such as flash management, RAID or erasure coding, access control, configuration, encryption, compression/deduplication, monitoring, and more. The interface for clients or hosts connecting to these storage systems often requires enhanced storage traffic capabilities over protocols like RDMA or TCP and storage virtualization. Traditionally, these comprehensive tasks are handled by general-purpose x86 CPUs, which consume more CPU resources as network speeds escalate and security demands intensify.
A notable shift in this landscape involves integrating specialized processing units within the storage servers and the client or host environments. In the client or host setups, these processors accelerate widely used storage protocols and manage encryption for data in transit. They also enable the virtualization of network storage, presenting it as high-performance local flash storage. This approach significantly reduces the CPU load for storage and networking virtualization tasks, allocating more CPU resources for core applications.
In storage architectures, whether monolithic or distributed, there's typically a set of front-end controllers that oversee access to the backend storage units. These advanced processors can replace traditional network cards, taking over tasks related to data movement, encryption, and RDMA from the CPU. This optimization allows the front-end controllers to either accommodate more users or execute additional storage software more efficiently. Within dedicated storage nodes, these processors can assume full responsibility for storage software and control functions, potentially replacing the need for x86 CPUs and network cards. This transition can considerably reduce the size, weight, and power requirements.
By leveraging technologies like NVMe over Fabrics (NVMe-oF) and GPUDirect Storage, alongside features for encryption, scalable storage, data integrity, decompression, and deduplication, these specialized processing units ensure high-performance access to storage with latencies competitive with direct-attached storage solutions. This evolution marks a significant step towards optimizing storage system performance while minimizing resource consumption and operational costs.
The NVIDIA BlueField-3 DPU (Data Processing Unit) represents significant advancements in data center infrastructure technology. It focuses on enhancing modern data centers' efficiency and capabilities, particularly in handling the demands of AI and Machine Learning workloads. As a third-generation DPU, BlueField-3 is designed to offload, accelerate, and isolate data center workloads, improving overall performance and security.
BlueField-3 offers an impressive 400 gigabits per second (Gb/s) Ethernet and InfiniBand connectivity, a considerable bandwidth leap that facilitates faster data transfer rates within data centers. This capability is critical for applications that require high-speed data processing and movement, such as AI, Machine Learning, and big data analytics.
One key feature of the BlueField-3 DPU is its integration with GPUDirect Storage technology. GPUDirect Storage allows Direct Memory Access (DMA) transfers between GPU memory and storage, bypassing the need to move data through the CPU. This direct path reduces latency, increases throughput, and lowers CPU utilization, which is particularly beneficial for AI and Machine Learning workloads that require fast access to large datasets.
In collaboration with NVIDIA, VAST Data has leveraged the BlueField-3 DPU to revolutionize data center architecture by developing a new infrastructure that significantly enhances parallel data services. VAST Data's approach involves using BlueField-3 DPUs as storage controllers within NVIDIA GPU servers. This configuration enables the creation of highly efficient and scalable storage solutions optimized for AI and Machine Learning applications. Integrating VAST Data storage technology with NVIDIA BlueField-3 DPUs allows up to 70% savings in power and space, demonstrating the potential for substantial improvements in data center efficiency and sustainability.
Furthermore, the combination of NVIDIA BlueField-3 DPUs and VAST Data's storage solutions facilitates the construction of AI clouds with a parallel system architecture. This setup enhances both storage and database processing, enabling more efficient handling of AI workloads and accelerating innovation in AI research and development.
Overall, NVIDIA's BlueField-3 DPU technology, in conjunction with GPUDirect Storage and VAST Data's innovative storage solutions, represents a significant step forward in the evolution of data center infrastructure. This powerful combination promises to transform data centers, making them more efficient, secure, and capable of meeting the demands of the AI era.
NVIDIA BlueField-3 DPU Component Side (Ref 3)
NVIDIA BlueField-3 DPU Print Side (Ref 4)
Table
Item 1
Interface: DPU SoC
Description: 8/16 Arm-Cores DPU SoC
Item 2
Interface: Networking Interface
Description: The network traffic is transmitted through the DPU QSFP112 connectors
The QSFP112 connectors allow the use of modules and optical and passive cable interconnect solutions
Item 3
Interface: Networking Ports LEDs Interface
Description: One bi-color I/O LEDs per port to indicate link and physical status
Item 4
Interface: PCI Express Interface
Description: PCIe Gen 5.0/4.0 through an x16 edge connector
Item 5
Interface: DDR5 SDRAM On-Board Memory
Description: 20 units of DDR5 SDRAM for a total of 32GB @ 5200 or 5600MT/s
128bit + 16bit ECC, solder-down memory
Item 6
Interface: NC-SI Management Interface
Description: NC-SI 20 pins BMC connectivity for remote management
Item 7
Interface: USB 4-pin RA Connector
Description: Used for OS image loading
Item 8
Interface: 1GbE OOB Management Interface
Description: 1GbE BASE-T OOB management interface
Item 9
Interface: MMCX RA PPS IN/OUT
Description: Allows PPS IN/OUT
Item 10
Interface: External PCIe Power Supply Connector
Description: An external 12V power connection through an 8-pin ATX connector
Applies to models: B3210E, B3210 and B3220
Item 11
Interface: Cabline CA-II Plus Connectors
Description: Two Cabline CA-II plus connectors are populated to allow connectivity to an additional PCIe x16 Auxiliary card
Applies to models: B3210E, B3210 and B3220
Item 12
Interface: Integrated BMC
Description: DPU BMC
Item 13
Interface: SSD Interface
Description: 128GB
Item 14
Interface: RTC Battery
Description: Battery holder for RTC
Item 15
Interface: eMMC
Description: x8 NAND flash
NVIDIA BlueField Fabric (Ref 5)
-Resources
Accelerating AI Storage Access with the NVIDIA BlueField DPU (Embedded Link)
NVIDIA BlueField-3 Networking Platform Datasheet (Embedded Link)
NVIDIA BlueField-3 Networking Platform Introduction Guide (Embedded Link)
NVIDIA BlueField-3 Networking Platform User Guide (Embedded Link)
NVIDIA BlueField-3 Networking Platform Administrator Quick Start Guide (Embedded Link)
Section 5 - The VAST Data Platform with NVIDIA BlueField-3 DPUs for Pediatric Disease Precision Medicine Approach Acceleration
The Genomics Secondary Analysis for Pediatric Disease Precision Medicine framework advances the AWS Genomics Secondary Analysis for Precision Medicine with the advanced features of the VAST Data Platform, leveraging the power of NVIDIA BlueField-3 DPUs to create a bespoke solution tailored for pediatric precision medicine. This enhanced system architecture integrates cutting-edge storage solutions, AI-accelerated analytics, and advanced data processing units, yielding a robust, scalable, and highly efficient platform for genomics data analysis.
System Architecture
This solution's core involves deploying the VAST Data Platform across NVIDIA GPU servers equipped with BlueField-3 DPUs. By integrating VAST's operating system directly onto BlueField-3 DPUs, we facilitate the seamless parallelization of storage and database processes at scale, which is essential for handling large-scale pediatric genomic datasets. This setup ensures:
Direct High-Speed Data Pathways: Utilizing GPUDirect Storage technology, genomics data can be moved directly between GPU memory and storage, bypassing traditional CPU bottlenecks, reducing latency, and enabling faster data access and analysis, which is essential for timely pediatric disease characterization and treatment strategy formulation.
Containerized Parallel Processing: State-of-the-art containerization on NVIDIA BlueField DPUs enables running VAST's parallel services operating system, integrating storage and database processing directly within AI servers. This allows for efficient linear scalability in data services, crucial for extended genomics analyses across diverse pediatric patient datasets.
Scalable and Secure Data Management: The combined infrastructure supports advanced analytics and Machine Learning for predictive modeling and ensures robust data integrity and security measures – a paramount consideration in handling sensitive pediatric genomics data.
Benefits of VAST Data Platform Genomics Secondary Analysis for Precision Medicine
While the AWS Genomics Secondary Analysis that I developed during my research at UC Berkeley provides a comprehensive cloud-based platform for genomics data analysis, the enhanced VAST Data Platform solution offers several distinct advantages:
Reduced Latency in Data Processing: Direct Memory Access facilitated by GPUDirect Storage and BlueField-3 DPUs significantly shortens the data path, enabling quicker data analysis, which is critical in rapid genomic sequencing and variant analysis for pediatric precision medicine.
Increased Efficiency and Scalability: The solution markedly increases data processing efficiency by deploying storage and database processes directly on BlueField-3 DPUs. This allows for the scalable and simultaneous analysis of extensive pediatric datasets that cloud-based solutions may struggle to process in real-time.
Enhanced Data Security: Pediatric genomics involves handling highly sensitive data, and the bespoke VAST data platform architecture offers superior data isolation and encryption capabilities, offering a higher level of security than traditional cloud environments.
Cost-Effectiveness at Scale: Leveraging VAST's power and space savings, alongside a significant reduction in CPU utilization due to offloading to DPUs, provides a cost-efficient solution, particularly at scale, which can be especially beneficial for large-scale pediatric genomics projects.
This approach enables a Genomics Secondary Analysis for Pediatric Disease Precision Medicine solution harnessing the unique capabilities of VAST Data Platform and NVIDIA BlueField-3 DPUs, offering a robust, efficient, and scalable architecture designed to meet the specific needs of precision medicine in pediatric care. Through enhanced data processing speed, scalability, and security, this solution positions research and clinical practices at the forefront of pediatric genomics, promoting rapid, informed, and personalized treatment pathways for children affected by genetic diseases.
Section 6 - Leveraging VAST Data Storage and Supermicro Hyper-Scale Servers with NVIDIA BlueField-3 DPUs for Advanced Healthcare and Life Sciences for Pediatric Cancer Precision Medicine Research
Solution Overview
The objective is to establish a robust and scalable software architecture that leverages the VAST Data Platform with Supermicro Hyper-Scale Servers alongside NVIDIA BlueField-3 DPUs to accelerate pediatric cancer research, mainly focusing on precision medicine applications. This architecture aims to integrate fastq.gz genomic files with patient phenotypic data and previous treatment outcomes to facilitate the development of targeted cancer treatments.
To achieve this, we propose an AI workflow utilizing a streamlined set of software tools to process genomic data, integrate clinical data, and implement Machine Learning models to predict disease outcomes and treatment responses. The selected software tools for this solution are Databricks Lakehouse, Apache Spark, Snowflake, and Kafka, which support data formats such as Parquet and Arrow and programming in Python for script automation and model training. The solution approach outlined below is only a partial solution, and it is not the only way to achieve the end goal. Other AI workflow software tools could be positioned to process genomic data, integrate clinical data, and implement Machine Learning models to predict disease outcomes and treatment responses.
Detailed Solution Process Workflow
1. Data Acquisition and Management
Ingest fastq.gz genomic files and patient phenotypic data into the VAST DataStore using Kafka for efficient data streaming.
Use VAST DataBase to organize and manage sequencing data, ensuring scalability and security.
2. Data Processing and Analysis
Deploy containerized bioinformatics tools (BWAMem, SAMTools, BCFTools) in the VAST DataEngine for initial genomic data processing. Output files in Parquet and/or Arrow formats for optimized performance in later analytics steps.
Utilize Apache Spark within the VAST DataEngine for distributed data processing, which can handle large-scale datasets efficiently.
3. Integration and Storage
Aggregate processed genomic data with patient phenotypic data and previous treatment outcomes in Snowflake, leveraging its secure, scalable data warehousing capabilities.
Store Variant Call Format (VCF) files and integrated clinical data in VAST DataBase for persistent, secure storage and easy retrieval.
4. Machine Learning and Analytics
Employ the Databricks Lakehouse platform for Machine Learning model development. This integrates seamlessly with Snowflake and VAST DataPlatform, offering a unified environment for data lake and warehouse capabilities.
Utilize Apache Spark for large-scale data analytics and train Machine Learning models using Python, with libraries such as TensorFlow or PyTorch, to predict disease outcomes or treatment responses.
5. Workflow Management and Automation
Manage the workflow from raw sequence data to analytics insights using Kafka for data movement and Spark Streaming for real-time data processing.
Automate repetitive tasks and streamline process flow with Python scripts, ensuring efficiency and minimizing manual intervention.
Benefits and Impact
This architecture leverages the high-performance capabilities of the VAST Data Platform with Supermicro Hyper-Scale Servers and NVIDIA BlueField-3 DPUs, providing a scalable, secure, and efficient solution for processing and analyzing pediatric cancer genomic data. By combining advanced data storage and processing technologies with intelligent Machine Learning models, this solution accelerates the development of precision medicine for pediatric cancer. It promises to enhance disease characterization and treatment strategy formulation, ultimately improving patient outcomes.