Over twelve years ago, I embarked on a journey to help accelerate innovation in precision medicine through the power of High-Performance Computing (HPC). During my 20+ year career, I’ve seen traditional HPC storage systems and parallel file architectures limit the speed and scalability needed for groundbreaking research. When I joined the VAST Data team, it was clear that the VAST Data Platform would be the key to overcoming these bottlenecks. With VAST, researchers can focus entirely on innovation, unburdened by the constraints of legacy systems.
Today, I want to share how Quantum Computing is poised to transform the development of preventative cancer therapies. I'll cover quantum simulations to model the molecular interactions of BRCA1 and BRCA2, genes crucial in breast cancer prevention. These simulations at the quantum level can uncover potential drug candidates by predicting how molecules interact with patient-specific genetic mutations. But the story doesn’t end there.
We can predict how drug compounds will behave across diverse patient genomes by integrating quantum results with HPC-powered AI training models on NVIDIA DGX H100 systems. This Accelerated Quantum Supercomputing approach enables the identification of high-potential gene therapies tailored to individual patients, marking a leap forward in personalized medicine.
With VAST Data's DASE (Disaggregated, Shared-Everything) Architecture, and the ability to scale seamlessly between Quantum and Classical HPC-AI Workloads, VAST enables researchers to turn these transformative possibilities into a reality. I will cover five major sections below; the content will take 25 minutes to read. Stick with me, and we'll get through this together.
Content Sections
Section 1 - BRCA1 and BRCA2 Gene Identification for Breast Cancer
Section 2 - Quantum Computing Overview
Section 3 - High-Density Quantum Controllers and Bridges
Section 4 - End-to-End Workflow for Drug Research Using Quantum and HPC
Section 5 - VAST Data Platform – The Optimal Enabler for Quantum-to-HPC Workflows
Section 1 - BRCA1 and BRCA2 Gene Identification for Breast Cancer
Breast Cancer Introduction
Breast cancer is one of the most common types of cancer in women. A complicated factor in breast tumor development is the tumor's direct proximity to lymph nodes and blood vessels. If a tumor is located in the center of the breast, then it may be more likely to spread to these regions than if it is on the periphery.
BRCA1 And BRCA2 Genes
BRCA1 and BRCA2 are two human DNA repair genes in the genome (Chromosomes 13 and 17), essential in preventing cancer formation. These genes work to recognize, repair, and replace damaged strands of DNA, which helps protect cells from developing genetic mutations that could lead to tumor growth. Mutations in either gene can be inherited from a parent or acquired due to environmental factors, and they increase one’s risk for certain types of tumors, such as breast, ovarian, and prostate cancers.
The function of BRCA1 and 2 lies in their ability to recognize structural problems within the genome, especially those caused by carcinogenic chemicals or other environmental exposures. They help to keep cell processes on track by identifying potential issues before they become dangerous and correcting them when necessary. In addition, these proteins play an indirect role in preventing cancer through their ability to protect against inflammation-related damage that can eventually lead to tumor formation.
Understanding how these genes work is essential for assessing personal risk for particular forms of cancer and targeting preventive strategies for those who may benefit from their use. These DNA repair proteins are crucial in keeping the human cell safe from harm caused by environmental agents and preventing genetic errors that could otherwise lead to tumor growth.
BRCA Genes Illustration (Copyright: Adam Jones)
Treatment Options for BRCA1 and/or BRCA2 Mutations
Individuals with BRCA1 and 2 gene mutations have several preventative treatment options depending on whether or not they have a family history of breast cancer.
For those with a family history of the disease, the most recommended preventive measure is increased screening for early signs of cancer. This may include regular mammograms, MRIs, or other imaging techniques. Additionally, some people who carry these gene mutations may opt to undergo prophylactic surgery to remove areas of tissue that could be at greater risk of developing tumors due to their genetic makeup.
Those without a family history of breast cancer can still benefit from preventative measures such as lifestyle modifications to reduce their risk. Such changes can include eating a healthy diet, exercising regularly, avoiding smoking, limiting alcohol consumption, and getting adequate sleep each night. In addition, many women choose to take medications such as Tamoxifen if they test positive for one or both of these genes to reduce their chance of tumor formation over time.
Ultimately, different treatment options exist for those with BRCA1 and 2 gene mutations depending on risk factors such as age and family history. Increased screening can identify potential issues before they become dangerous, while prophylactic surgery can remove areas at risk for tumor growth; however, even those without a family history should look into lifestyle modifications and medications that are proven to reduce one’s chance of developing certain types of cancers.
Treatment Approach for this Article
We will explore how Accelerated Quantum Supercomputing can revolutionize breast cancer prevention by leveraging Quantum Computing to simulate BRCA1 and BRCA2 at the molecular level. These simulations have the continued potential to uncover critical insights into how mutations impact protein function and interaction. By coupling quantum-generated data with HPC-driven AI models, this approach enables the creation of personalized precision therapies tailored to individual genetic profiles.
Section 2 - Quantum Computing Overview
Quantum Computing Introduction
Quantum computing is a revolutionary approach to computation that leverages the principles of quantum mechanics to process data in ways far beyond the capabilities of classical systems. Unlike classical computers, which process information in binary (0s and 1s), quantum computers use qubits, capable of existing in multiple states simultaneously due to the principles of superposition and entanglement.
This capability allows quantum computers to solve problems involving vast datasets and intricate interactions, offering opportunities in fields such as drug discovery, artificial intelligence, and material sciences. By harnessing the immense scalability and speed of quantum mechanics, quantum computing opens new frontiers in areas where classical systems are limited.
Below, I've included a photo of most individual's first view of a Quantum Computer. It is the Quantum Cryostat/Dilution Refrigerator. Long story short, Quantum Computers are highly sensitive to environmental noise, including heat, which can disrupt their fragile quantum states—a phenomenon known as decoherence. The cryostat allows the Quantum Computer to maintain coherence and perform reliable quantum computations by reducing thermal energy to near-zero levels.
Quantum Cryostat/Dilution Refrigerator Photo (Copyright: Adam Jones)
Classical vs. Quantum Computing
In classical computing, information is processed using bits that represent either "on" (1) or "off" (0). Logical operations manipulate these bits sequentially to solve problems. Quantum computing, however, leverages qubits, which can exist in multiple states simultaneously. This capability drastically increases computational power by allowing quantum computers to explore multiple solutions at once.
An Analogy: Imagine a straight, one-dimensional line for classical computing, where solutions are found by moving point-by-point. In contrast, quantum computing resembles a three-dimensional sphere, where every point represents a potential solution. This expanded possibility space enables quantum systems to tackle problems that classical computers cannot feasibly solve.
Quantum Mechanics and Qubits
Quantum mechanics provides the foundation for quantum computing by explaining how particles behave at microscopic scales. Concepts like wave-particle duality, quantum tunneling, and entanglement govern the behavior of qubits.
Qubits, the fundamental units of quantum computation, are created using various technologies, such as:
Trapped Ions: Atoms suspended in a vacuum and manipulated using electric and magnetic fields.
Superconducting Circuits: Microelectronic components like Josephson junctions, which operate at cryogenic temperatures to maintain quantum states.
These qubits are manipulated using precise microwave signals to perform calculations, enabling tasks that outperform classical systems in specific scenarios.
Qubit Superposition Overview
Superposition is a key property of qubits that allows them to exist in multiple states simultaneously. Unlike a classical bit, which must be either 0 or 1, a qubit can represent 0, 1, or any combination of both at the same time. This property enables quantum computers to process vast amounts of information in parallel, significantly accelerating problem-solving for complex scenarios.
For example, superposition allows a quantum computer to evaluate millions of potential solutions simultaneously during a simulation, making it particularly valuable in molecular modeling.
Classical BIT to QUBIT Comparison Illustration (Copyright: Adam Jones)
Qubit Entanglement Overview
Entanglement is another critical phenomenon in quantum computing, where two or more qubits become correlated in such a way that the state of one qubit is intrinsically linked to the state of another, regardless of the distance between them. When qubits are entangled, the measurement of one qubit’s state instantly determines the state of the other.
Entanglement enables quantum computers to perform complex computations with fewer qubits. This interconnected behavior allows for faster information processing and more efficient solutions to problems that involve interdependent variables, such as optimizing molecular interactions in drug design.
Quantum Computing Hardware Overview
A quantum computer relies on several highly specialized components to maintain and utilize the delicate quantum states of qubits.
1. Cryostat/Dilution Refrigerator
Often resembling a gold-plated chandelier, this device cools the quantum processor to near absolute zero (around 0.01 Kelvin or -459°F). At these temperatures, qubits maintain their coherence, unaffected by thermal noise or environmental factors.
Gold-Plated Components: These are used to ensure thermal conductivity and reduce electromagnetic interference, protecting the qubits.
Thermal Layers and Microwave Wiring: Successive cooling stages progressively lower temperatures, while specialized wiring delivers precise signals to control the qubits.
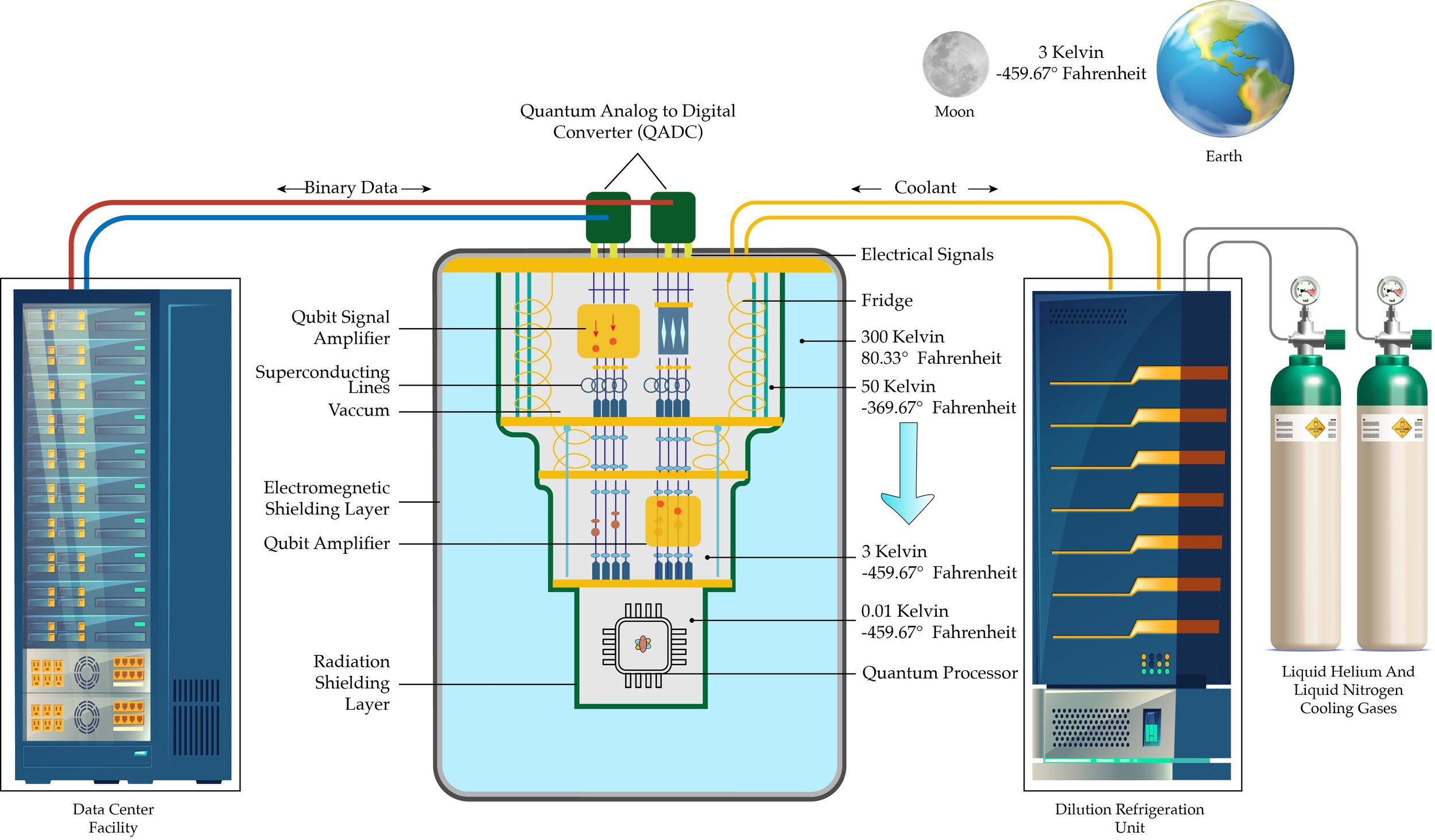
Quantum Computer Hardware Architecture Illustration (Copyright: Adam Jones)
2. Quantum Processor (QPU)
Located at the base of the cryostat, the QPU houses the qubits that store and process quantum information. Superconducting lines connect these qubits to external systems, ensuring low-resistance signal transmission.
3. Quantum Analog-to-Digital Converters (QADCs)
These devices convert analog quantum signals into classical data formats, enabling seamless communication between quantum and classical computing systems.
Quantum Computer to Quantum Analog-to-Digital Converter (QADC) Process Flow Illustration (Copyright: Adam Jones)
4. Signal Amplifiers and Superconducting Lines
Amplifiers enhance the weak signals emitted by qubits, ensuring accurate data readout for processing.
The Challenge of Decoherence
Qubits are highly sensitive and can lose their quantum state through interactions with their environment, a phenomenon known as decoherence. Factors like thermal energy, electromagnetic interference, or particle collisions can disturb their state, rendering them unusable for computations. To prevent this, the cryostat maintains the quantum processor at ultra-low temperatures, preserving the stability and functionality of qubits.
Section 3 - High-Density Quantum Controllers and Bridges
QM OPX1000 High-Density Quantum Controller Introduction
The QM OPX1000 High-Density Quantum Controller is designed to manage and control qubits within a quantum computing system. It features multiple analog and digital input/output channels, enabling it to interface directly with quantum processors and associated hardware. It interfaces directly with the quantum processor, controlling qubits by generating precise microwave and low-frequency signals necessary for qubit manipulation and measurement. The QM OPX1000 High-Density Quantum Controller is designed to interface seamlessly with both quantum processors and classical computing systems, facilitating efficient quantum operations. The various channels connect within a quantum computing setup as outlined below.
Analog Output Channels: These channels generate precise control signals, such as microwave pulses, to manipulate qubits within the quantum processor. The analog outputs are connected directly to the quantum processor's control lines, delivering the necessary signals to perform quantum gate operations and qubit state manipulations.
Analog Input Channels: Analog input channels receive signals from the quantum processor, typically carrying information about qubit states after measurement operations. These input channels are connected to the readout circuitry of the quantum processor, capturing analog signals that represent the qubits' measured states. The OPX1000 then digitizes these signals for further processing.
Digital Input/Output (I/O) Channels: The digital I/O channels facilitate communication between the OPX1000 and classical computing components. They are used for tasks such as synchronization, triggering events, and exchanging control signals with classical hardware. These channels connect to various classical devices and systems that coordinate with the quantum processor, ensuring coherent operation across the quantum-classical interface.
Video Resource: The OPX1000 Quantum Control Platform Walkthrough Video on YouTube lasts 7 minutes and 36 seconds and provides a good OPX1000 Overview.
QM OPX1000 High-Density Quantum Controller Hardware Overview
The QM OPX1000 can accommodate eight modules per chassis, providing the channel capabilities listed below.
64 Analog Output Channels
16 Analog Input Channels
64 Digital Input/Output Channels
(Ref 1) - QM OPX1000 Chassis Photo
(Ref 2) - QM OPX1000 Front-End Modules Photo
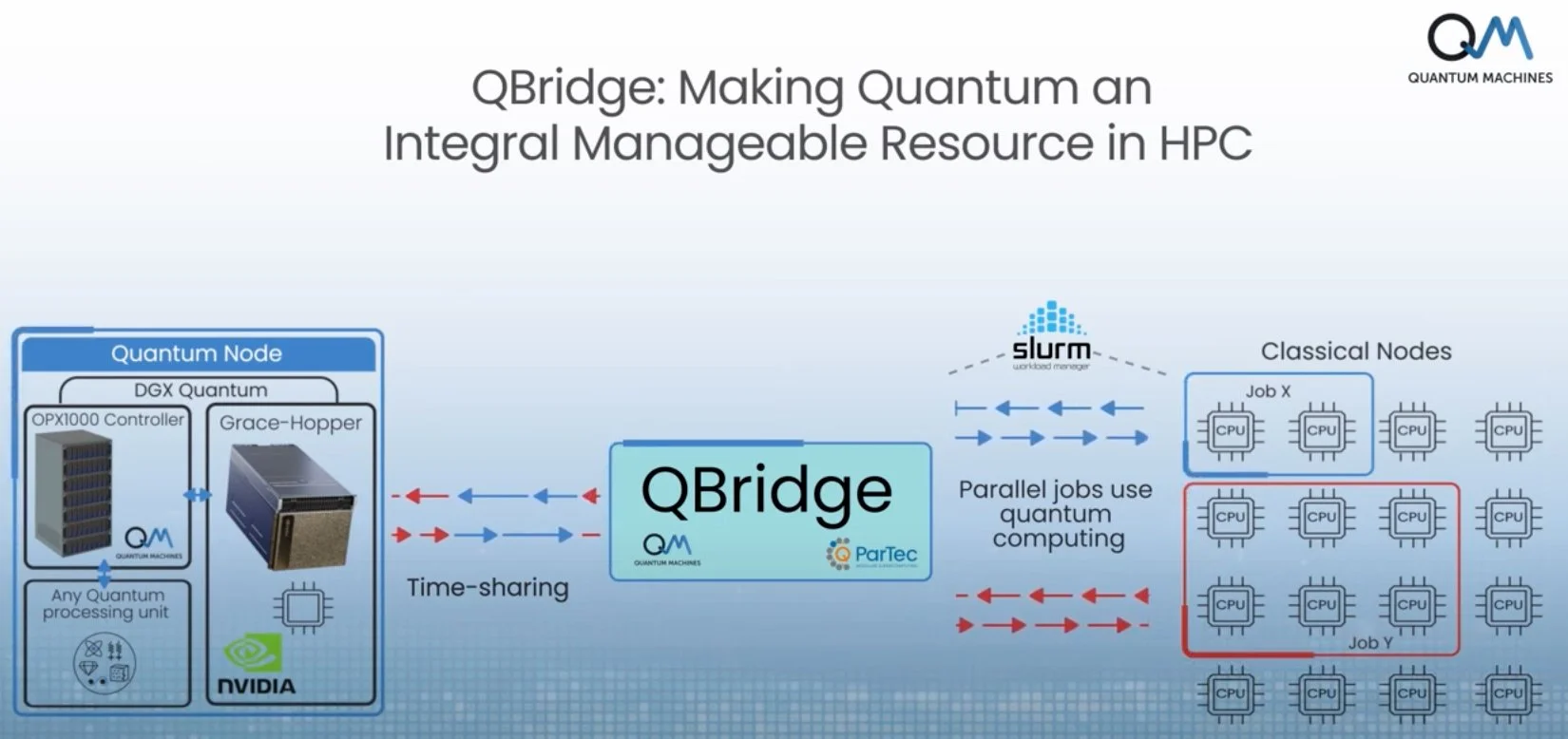
QBridge Introduction
QBridge is a universal software solution co-developed by ParTec and Quantum Machines to integrate quantum computers into HPC environments seamlessly. It enables multiple HPC users to execute hybrid workflows across classical and quantum computing resources, facilitating efficient co-scheduling and resource management.
In the context of a quantum computing setup involving the QM OPX1000 High-Density Quantum Controller and an NVIDIA DGX H100 system, QBridge plays a pivotal role by:
Coordinating Resources: QBridge manages the allocation and scheduling of tasks between the quantum processor controlled by the OPX1000 and the classical computing resources of the DGX H100, ensuring optimal utilization of both systems.
Facilitating Hybrid Workflows: By enabling seamless communication between quantum and classical components, QBridge allows complex computations to be divided appropriately, leveraging the strengths of each platform.
Enhancing User Accessibility: QBridge provides a secure and efficient environment for HPC centers, cloud providers, and research groups to integrate quantum computing into their existing infrastructure, making advanced computational resources more accessible.
Qbridge Diagram
By incorporating QBridge into these designs, organizations can harness the combined power of quantum and classical computing, accelerating innovation in Healthcare and Life Sciences.
Section 4 - End-to-End Workflow for Drug Research Using Quantum and HPC
In this section, we will explore the workflows for personalized therapies for specific genes. The entire system—quantum computing, GPUs, and HPC—works together step by step as outlined below.
Step 1: Molecular Data Collection
1. Genomic Data Analysis
Collect genomic data from patient DNA samples.
Sequence the regions of interest, focusing on Chromosome 17 (BRCA 1) and Chromosome 13 (BRCA 2).
Use computational tools (like GATK, and BWA-MEM) to map and identify mutations in these genes.
Additional Resources: Directly below is the content I created for my Primary and Secondary Genomics Analysis for Variant Call Format (VCF) Solution during my UC Berkeley post-graduate studies.
2. Protein Structures
Extract the 3D structures of BRCA 1 and BRCA 2 proteins from databases like Protein Data Bank (PDB).
Focus on mutated regions to identify structural or functional defects.
Step 2: Quantum Computing for Molecular Simulation
1. Setup Quantum Simulations
Load the 3D molecular structures of BRCA 1 and BRCA 2 proteins into the Quantum Processing Unit (QPU) controlled by the QM OPX1000.
Simulate molecular interactions at the quantum level.
How candidate molecules (e.g., gene-editing tools, inhibitors) bind to the mutated proteins.
Explore potential conformational changes that restore protein function or prevent harmful interactions.
2. Quantum Advantages
The QPU analyzes the quantum states of molecular components, exploring millions of possible interactions simultaneously.
For example, it can determine the optimal configuration of a drug molecule to block harmful genetic mutations.
3. Error Correction & Data Processing
The OPX1000 ensures accuracy by correcting any errors during the quantum computation.
Results are sent as quantum data to the QBridge, which translates the quantum output into a format the classical system can process.
Step 3: Data Transition to HPC AI
1. Data Processing via QBridge
Results from quantum simulations are sent to the DGX H100 system via QBridge.
QBridge manages data formatting and workload orchestration between the quantum and classical systems.
Step 4: GPU-Accelerated AI for Personalized Insights
1. Advanced AI Model Training
The DGX H100 uses the molecular interaction data to train AI models.
These models predict how the drug compound will behave across different patient genomes or specific genetic variations.
Identify high-potential molecules for gene therapy.
Rank molecules based on binding affinity, specificity, and stability.
2. Patient-Specific Therapy Design
AI algorithms simulate therapy outcomes on a patient-by-patient basis. For instance:
How effective a drug will be in blocking a harmful mutation in a specific patient's DNA.
Small Molecule Inhibitors: Create molecules that enhance the mutated protein’s function or inhibit pathways leading to cancer.
Whether side effects are likely based on the patient's unique genetic makeup.
Step 5: HPC Integration for Population-Level Studies
1. Scale to Larger Studies
Using HPC resources, you analyze the AI results across large patient datasets.
This step helps identify patterns or optimize therapies for broader genetic variations within a population.
Validate personalized therapies by comparing patient-specific results against population-level genomic variations.
2. Collaborative Research
Data is stored and shared efficiently using the VAST Data Platform.
VAST’s architecture ensures researchers worldwide can access, analyze, and collaborate on this data in real-time.
AI models simulate the long-term effects and success rates of the developed therapies.
Generate predictive outcomes for different demographic and genetic backgrounds.
Step 6: Final Outputs and Clinical Translation
1. Clinical Trials
Use insights from quantum-classical workflows to identify the most promising therapies.
Design trials to test these therapies on a diverse cohort of patients.
2. Preventative Measures
Develop therapeutic protocols to prevent breast cancer in individuals identified as high-risk through genetic screening.
Therapies could include targeted molecular treatments.
As more data is collected from treatments, the system learns and improves, enabling faster and more precise therapies in the future.
Section 5 - VAST Data Platform – The Optimal Enabler for Quantum-to-HPC Workflows
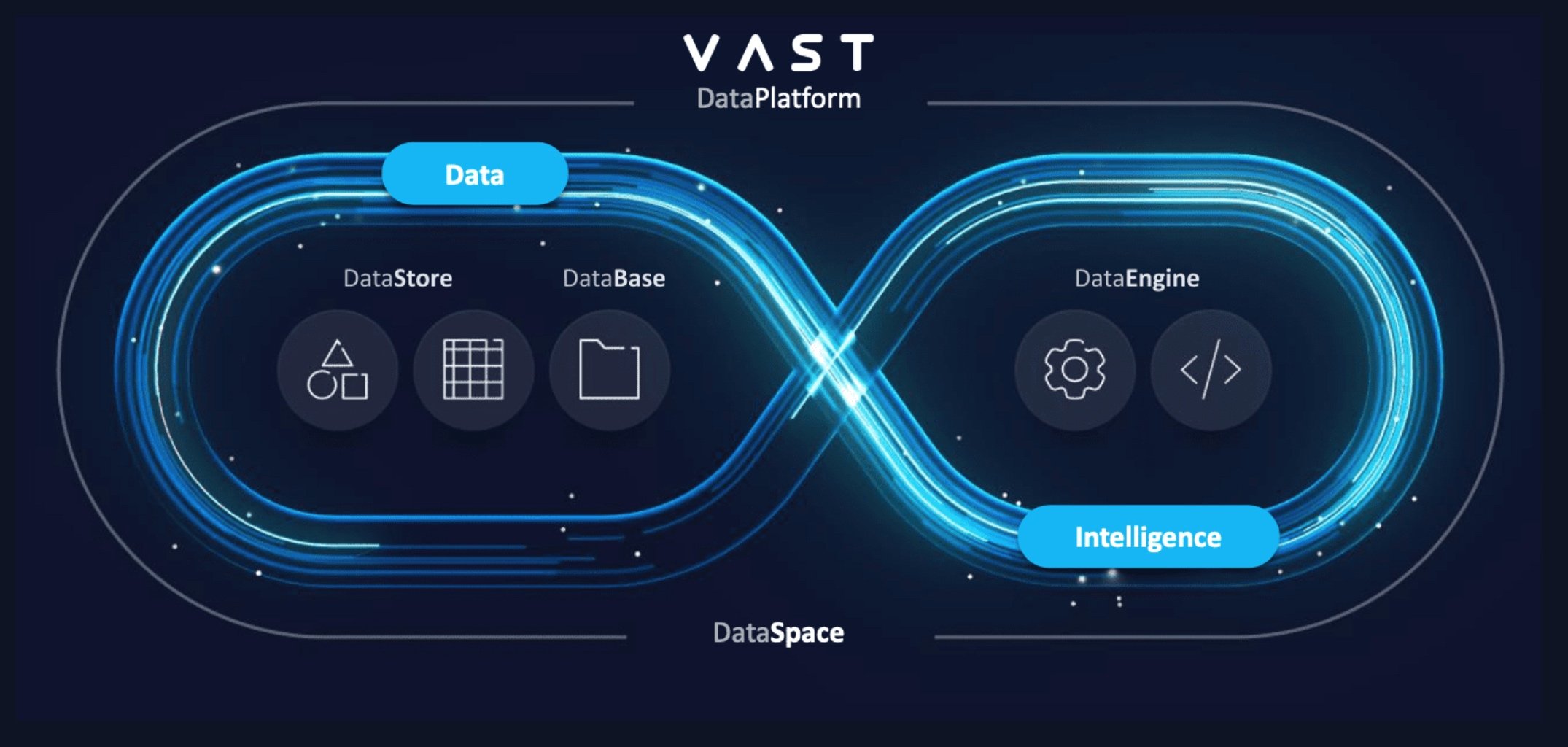
Why the VAST Data Platform for Quantum-to-HPC Workflows
The VAST Data Platform provides a transformative advantage by enabling seamless interaction between quantum computing, GPU-accelerated AI, and HPC systems. By addressing critical challenges inherent in traditional storage and workflow integration, VAST ensures that the entire end-to-end drug research workflow—from quantum simulations to AI-driven precision therapies—is faster, more efficient, and scalable. This is accomplished through the features below.
1. Disaggregated, Shared-Everything (DASE) Architecture: Simplifying Quantum-HPC Integration
Independent Scaling for Quantum and HPC Workloads
Quantum systems like the QM OPX1000 and classical AI systems like the NVIDIA DGX H100 have varying performance and capacity needs. The DASE architecture allows compute and storage to scale independently, ensuring neither system is constrained by the other.
For example, when quantum simulations generate large data sets, VAST’s DASE enables immediate access to this data for HPC AI processing without additional data migrations.
Data Consistency Across Quantum and Classical Systems
Traditional shared-nothing architectures struggle with synchronizing data across heterogeneous environments. VAST’s global namespace ensures quantum outputs are consistently and instantly accessible to AI and HPC nodes, eliminating delays.
2. Accelerating Workflow Management with Unified Data Access
Real-Time Data Availability
The VAST Data Platform consolidates scratch, nearline, and archive storage tiers into a single all-flash architecture. This eliminates the need for time-consuming data migrations, ensuring quantum simulation results are instantly accessible to GPU-driven AI models.
Simplified Data Orchestration
While tools like QBridge handle data formatting and workload orchestration, VAST ensures the underlying data infrastructure operates seamlessly. With native support for NFS, SMB, and S3 protocols, data flows between quantum and classical systems are efficient and require minimal configuration.
3. All-Flash Performance for Quantum Workflows
Handling Quantum Data Volumes
Quantum simulations produce vast datasets with complex metadata. VAST’s all-flash architecture ensures that these datasets are ingested, processed, and analyzed at lightning speed.
This allows researchers to iterate on molecular simulations quickly, refining potential drug candidates without storage bottlenecks.
Cost Efficiency at Scale
Unlike hybrid storage systems that combine SSDs and HDDs, VAST’s all-flash platform delivers superior performance at a comparable or lower cost. This affordability enables researchers to consolidate all workflow stages—quantum, AI, and HPC—into a single system.
4. Enhanced Collaboration and Scalability
Global Namespace for Collaborative Research
With the VAST Data Platform, researchers across the globe can access shared datasets in real-time. This capability fosters collaboration between quantum research teams and AI modeling groups, ensuring synchronized progress.
Scalability for Population-Level Analysis
As the system scales from individual patient data to population-wide genomic studies, VAST’s ability to handle exabyte-scale data ensures no compromise in performance or accessibility.
5. Advanced Data Protection for Critical Research
Erasure Coding for High Resilience
VAST’s erasure coding protects quantum and classical data without the overhead of traditional RAID systems. This ensures that critical quantum simulation results and AI-generated insights are safeguarded against data loss, even in large-scale environments.
Fault Tolerance in Continuous Workflows
With a focus on minimizing downtime, VAST ensures uninterrupted data availability during system expansions or maintenance, a critical requirement for time-sensitive drug research workflows.
6. Optimized for Modern Workloads
Quantum, AI, and HPC Harmony
VAST is designed for hybrid workloads, enabling quantum simulations, AI model training, and HPC-scale analysis to operate concurrently without performance degradation.
This optimization allows researchers to explore multiple therapeutic strategies in parallel, significantly reducing the time required to identify viable solutions.
Flexibility for Evolving Needs
As drug research workflows evolve, VAST’s architecture adapts to new demands, whether through increased quantum simulation complexity or expanding HPC capabilities for broader population studies.
7. How VAST Outperforms Legacy Systems
No Data Bottlenecks: Traditional systems often require manual data migrations and tuning between quantum, AI, and HPC stages. VAST eliminates these inefficiencies, delivering instantaneous data access.
Unified Platform: By consolidating all storage tiers and supporting diverse protocols, VAST reduces operational complexity, freeing researchers to focus on innovation rather than infrastructure management.
Scalable Precision Medicine: VAST’s combination of speed, scalability, and cost efficiency empowers researchers to expand precision medicine approaches from patient-specific therapies to population-wide preventive measures.
Final Thoughts: Enabling the Future of Drug Discovery
VAST Data allows experts to focus on using data versus managing it for things like proactively treating diseases! By seamlessly connecting quantum simulation data with AI-driven insights and HPC analysis, VAST accelerates the path from discovery to therapeutic application. Whether designing personalized small molecules or conducting population-level studies, VAST provides the foundational infrastructure that makes these possibilities a reality, paving the way for breakthroughs in Healthcare and Life Sciences.